Abstract
Background: PIM1 is a somatic hypermutation gene in diffuse large B-cell lymphoma (DLBCL) and its inhibitors exhibit a great value in application in multiple types of lymphoma. Nevertheless, investigation on its genetic alterations, biological characteristics, clinical significance and response to drugs is still lacking.
Methods: We integrated the genome sequencing (discovery cohort, n=162; validation cohort, n=1001) and transcriptome sequencing (discovery cohort, n=140; validation cohort, n=928) to capture more detailed insights into the potentially biological functions of PIM1 genetic alterations, and analyzed their relationship with biological characteristics and clinical value which provide a possibility for risk stratification and therapeutic exploitation for patients with DLBCL.
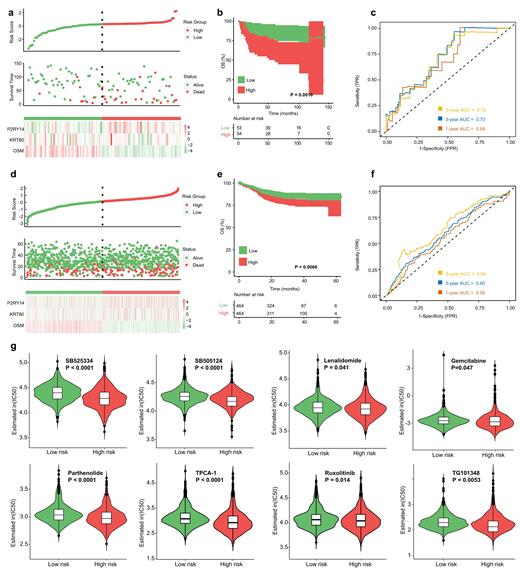
Results: PIM1 mutations were identified in 28.4% of DLBCL patients and significantly correlated with higher IPI scores (P=0.013), disease relapse (P=0.031) and CNS and/or testis involvement (P=0.001), as well as inferior PFS (P=0.022) and OS (P=0.0022). Multivariate analysis revealed that PIM1 mutation status was an independent poor prognostic factor (HR=2.86; 95% CI, 1.40-5.84; P=0.004). The most frequent PIM1 mutation type was missense mutations (84.1%), followed by frameshift deletions and nonsense mutations. During the distribution of base substitutions, C > T base substitution was predominant mutation type (54.4%), followed by C >G transversion (29.3%). During different exons, exon 4 of PIM1 was most often mutated. PIM1 mutations significantly co-occurred with the mutations of SETD1B (P<0.001) and CD79B (P=0.001), and was mutually exclusive to the SPEN mutation (P=0.024). We also explored the relationship between PIM1 mutations and genes distributed on previously reported signaling pathways in DLBCL and uncovered that mutations in MYD88 (P<0.001) and PRDM1 (P<0.001) involved in the NF-κB pathway were significantly enriched in the patients with PIM1 mutations. Unsurprisingly, patients with PIM1 mutations exhibited higher mutation frequencies in CD79B (P=0.001) involved in BCR pathway. PIM1 mutations were involved in immunoglobulin-related immune response, complement activation, B cell mediated immunity, B cell activation, antigen binding, and cytokine activity, which contributed to the signaling pathways of tumor microenvironment (e.g. cytokine-cytokine receptor interaction, chemokine signaling pathway, IL-17 signaling pathway, TNF signaling pathway), JAK-STAT and NF-κB. By MCODE analysis, five significant modules were obtained from the protein-protein interaction (PPI) network. Finally, a PIM1 mutation-related gene signature consisting of some independent prognostic factors was developed. According to the risk score, patients with high-risk score exhibited significantly shorter OS (P=0.0016) and PFS (P<0.001). The areas under the curve (AUCs) for the predictions of 1-, 3-, and 5-year OS were respectively 0.69, 0.70, and 0.72, suggesting that the risk score based on the PIM1 mutation-related gene signature had satisfactory sensitivity and specificity. We further found that compared to patients with low-risk score, patients with high-risk score had higher sensitivity to some drugs of targeting the immune microenvironment, including TGFβ receptor inhibitors SB525334 (P<0.0001) and SB505124 (P<0.0001) and immunomodulator Lenalidomide (P=0.041), as well as NF-κB inhibitors Parthenolide (P<0.0001) and TPCA-1 (P<0.0001) and JAK inhibitors Ruxolitinib (P=0.014) and TG101348 (P=0.0053), accompanying with significantly lower IC50 values. In addition, another common chemotherapeutic drug Gemcitabine was also predicted to be more sensitive for patients with high-risk score (P=0.047). Other targeted drugs such as Aurora kinase inhibitors VX-680 (P<0.0001) and ZM-447439 (P=0.014), Bcl-2 inhibitors Obatoclax Mesylate (P=0.00036) and Navitoclax (P<0.0001), and CDK inhibitors Roscovitine (P=0.0012), AT-7519 (P=0.0033), PHA-793887 (P<0.0001) and THZ2-49 (P=0.0053) also exhibited higher drug sensitivity for patients with high-risk score.
Conclusions: PIM1 mutations play a vital role in patient risk stratification and provide novel insights into therapeutic decision making for DLBCL patients with high-risk score.
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal